from math import fabs
from math import sqrt
from compas.geometry import length_vector
from compas_cem.diagrams import TopologyDiagram
from compas_cem.elements import Node
from compas_cem.elements import TrailEdge
from compas_cem.elements import DeviationEdge
from compas_cem.loads import NodeLoad
from compas_cem.supports import NodeSupport
from compas_cem.equilibrium import static_equilibrium
from compas_cem.plotters import TopologyPlotter
from compas_cem.plotters import FormPlotter
from compas_cem.optimization import Optimizer
from compas_cem.optimization import TrailEdgeForceConstraint
from compas_cem.optimization import DeviationEdgeParameter
# global controls
OPTIMIZE = True
PLOT = True
width = 4
height = width / 2
# Topology diagram
topology = TopologyDiagram()
# add nodes
topology.add_node(Node(1, [-width / 2, height, 0.0]))
topology.add_node(Node(2, [width / 2, height, 0.0]))
topology.add_node(Node(3, [0.0, height / 2, 0.0]))
topology.add_node(Node(4, [0.0, 0.0, 0.0]))
# add edges with negative values for a compression-only structure
topology.add_edge(TrailEdge(3, 4, length=-height/2))
topology.add_edge(DeviationEdge(1, 3, force=-sqrt(4.0)))
topology.add_edge(DeviationEdge(2, 3, force=-sqrt(2.0)))
topology.add_edge(DeviationEdge(1, 2, force=2.0))
# add supports
topology.add_support(NodeSupport(4))
# add loads
topology.add_load(NodeLoad(1, [0.0, -1.0, 0.0]))
topology.add_load(NodeLoad(2, [0.0, -1.0, 0.0]))
# auto generate trails and auxiliary trails
topology.build_trails(auxiliary_trails=True)
# form-finding
form = static_equilibrium(topology, eta=1e-5, tmax=100)
if OPTIMIZE:
# create optimizer
optimizer = Optimizer()
# add goal constraints
for edge in topology.auxiliary_trail_edges():
optimizer.add_constraint(TrailEdgeForceConstraint(edge, force=0.0))
optimizer.add_constraint(TrailEdgeForceConstraint(edge, force=0.0))
# add parameters
optimizer.add_parameter(DeviationEdgeParameter((1, 2), 1.0, 10.0))
optimizer.add_parameter(DeviationEdgeParameter((1, 3), 1.0, 10.0))
optimizer.add_parameter(DeviationEdgeParameter((2, 3), 10.0, 1.0))
# optimize
form = optimizer.solve_nlopt(topology, "SLSQP", 100, 1e-6)
# print out value of the objective function, should be a small number
print("Total value of the objective function: {}".format(optimizer.penalty))
print("Norm of the gradient of the objective function: {}".format(optimizer.gradient_norm))
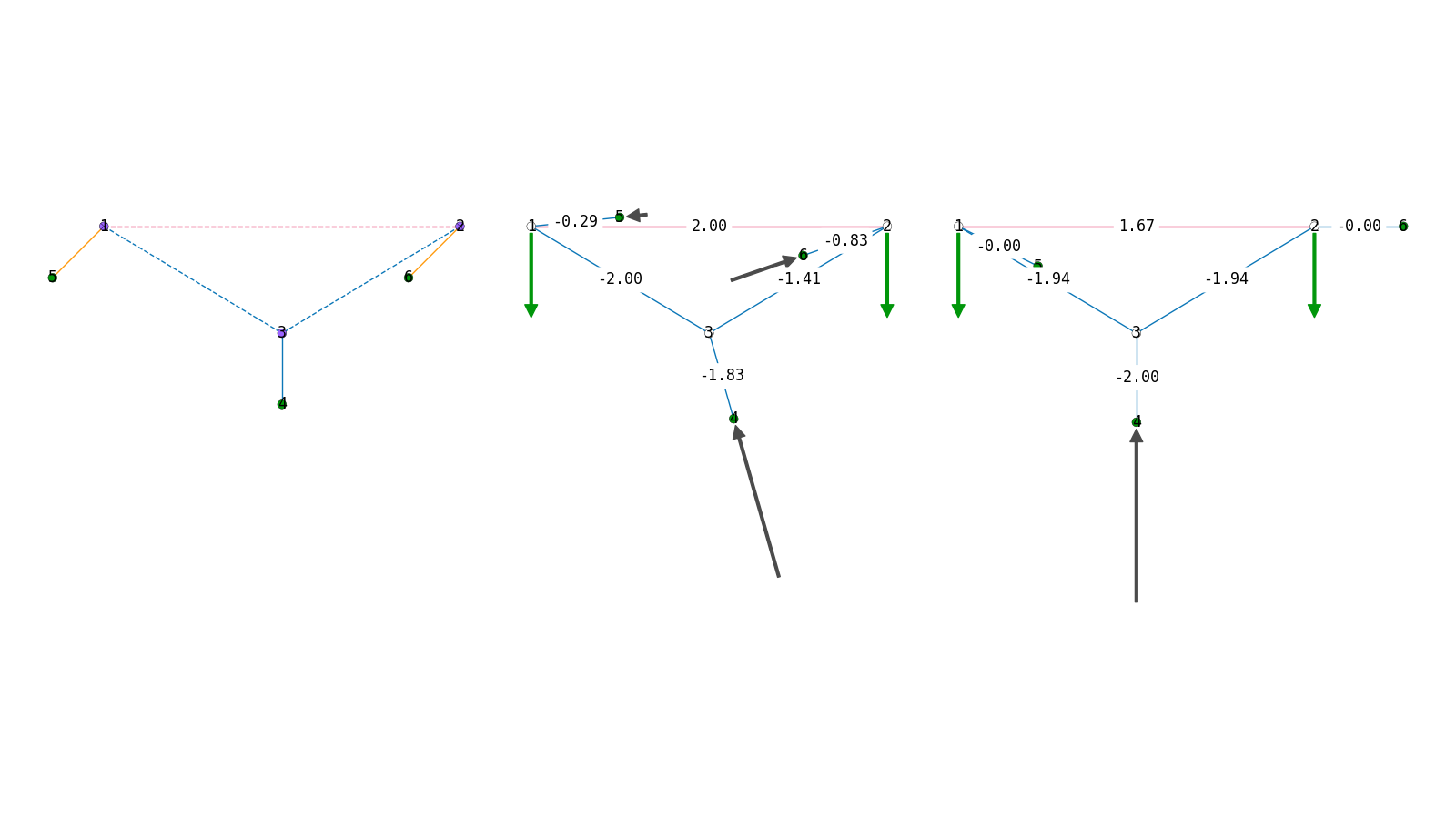
if PLOT:
plotter = TopologyPlotter(topology, figsize=(16, 9))
plotter.draw_nodes(radius=0.05, text="key")
plotter.draw_loads(radius=0.05, draw_arrows=True, scale=0.5)
plotter.draw_edges()
plotter.show()
plotter = FormPlotter(form, figsize=(16, 9))
keys = []
for node in form.nodes():
if form.is_node_support(node):
if length_vector(form.reaction_force(node)) <= 0.001:
continue
keys.append(node)
# keys = None
plotter.draw_nodes(keys=keys, radius=0.05, text="key-xyz")
plotter.draw_loads(keys=keys, scale=0.5)
plotter.draw_reactions(keys=keys, scale=0.5)
keys = [edge for edge in form.edges() if fabs(form.edge_force(edge)) >= 0.001]
# keys = None
plotter.draw_edges(keys=keys, text="force-length")
plotter.show()